Main objective of the project: Automate the process of detecting COVID-19 cases from chest radiograph images, using convolutional neural networks (CNN) through deep learning techniques. The complete project can be accessed here
Steps to reach the goal:
1- Data pre-processing
2- Model training and exposure of results
Step 2 - Model training and exposure of results
Databases used:
- X-ray images and chest CT scans of individuals infected with COVID-19 (COHE; MORRISON; DAO, 2020): link
- Images of lungs of individuals without any infection (KERMANY; ZHANG; GOLDBAUM, 2018): link
Packages used:
- Pandas
- Os
- PIL
- Tensorflow
- Sklearn
- Imutils
- Matplotlib
- Numpy
- Argparse
- Cv2
- Seaborn
Code used in the project:
The notebook with all the codes used in this step is available here
Note: the numbering and title of each step described in this tutorial correspond with the numbering and title contained in the notebook.
Steps to be followed:
1º Step – Import the libraries to be used
2º Step – Load the arrays built in the data pre-processing step and normalize the input data
3º Step – Split data into training data and test data
4º Step – Determining the architecture of the model to be trained
5º Step – Determine the hyperparameters and compile the model
6º Step – Train the model
7º Step – Observe the accuracy of the model and the loss function
8º Step – Observe which images the model hit
9º Step – Build a confusion matrix
Tutorial 2:
1º Step
Import the libraries to be used
We import the Tensorflow, Sklearn, Imutils, Matplotlib, Numpy, Argparse, Cv2, Os, Pandas and Seaborn libraries, since we will rely on them to carry out the training of the model referring to COVID-19 and the analysis of the results.
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.layers import Dropout
from tensorflow.keras.layers import MaxPooling2D
from tensorflow.keras.layers import Conv2D
from tensorflow.keras.layers import Flatten
from tensorflow.keras.layers import Input
from tensorflow.keras.layers import AveragePooling2D
from tensorflow.keras.layers import Dense
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.models import Model
from tensorflow.keras.applications import VGG16
from sklearn.preprocessing import LabelBinarizer
from tensorflow.keras.utils import to_categorical
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report
from sklearn.model_selection import train_test_split
from imutils import paths
import matplotlib.pyplot as plt
from sklearn.metrics import confusion_matrix
import argparse
import pandas as pd
import cv2
import numpy as np
import os
import seaborn as sn
%matplotlib inline
Note: some libraries have not been imported completely, such as, for example, Tensorflow, as we will not use all the functions contained therein. In this way, it facilitates the use of the library and the processing of codes/data.
2º Step
Load the arrays built in the data pre-processing step and normalize the input data
The “X_Train” and “Y_Train” arrays built in Step 1 were loaded and associated, respectively, with the variables “X_train” and “Y_train”. In addition, the variable X_train has been normalized for values ranging from 0 to 1.
X_train = np.load("/content/drive/My Drive/Python/COVID/Arrays/X_Train.npy")
X_train = X_train/255
Y_train = np.load("/content/drive/My Drive/Python/COVID/Arrays/Y_Train.npy")
3º Step
Split data into training data and test data
20% of the data referring to the images were separated for the model test. The function below returns four values that have been associated with four variables, namely: “X_train”, “X_test”, “Y_train” and “Y_test”.
It is possible to observe below that the number of cases is the same for the training dataset (n = 117) and also for the test (n = 30).
X_train,X_test,Y_train,Y_test = train_test_split(X_train,Y_train, test_size = 0.2, random_state = 40)
print(f"X_train shape: {X_train.shape} Y_train shape {Y_train.shape}")
print(f"X_test shape: {X_test.shape} Y_test shape {Y_test.shape}")
X_train shape: (117, 237, 237, 3) Y_train shape (117, 1)
X_test shape: (30, 237, 237, 3) Y_test shape (30, 1)
Note: the “random_state” parameter makes the random selection of images the same every time the function is executed.
4º Step
Determining the architecture of the model to be trained
The weights of the VGG16 architecture were loaded from the “imagenet” dataset, disregarding the top of the network. In addition, the input was defined with the size of the images in the image bank that we will use, namely: 237 x 237px and 3 color channels as depth. This information was associated with the “bModel” variable.
In addition, the architecture of the top of the network was determined, since the top of the network was removed from the “imagenet” dataset. This architecture was associated with the “tModel” variable.
Finally, the “bModel” and “tModel” variables were merged into the “model” variable. This last variable represents the model that will be trained.
bModel = VGG16(weights="imagenet", include_top=False,
input_tensor=Input(shape=(237, 237, 3)))
tModel = bModel.output
tModel = AveragePooling2D(pool_size=(4, 4))(tModel)
tModel = Flatten(name="flatten")(tModel)
tModel = Dense(64, activation="relu")(tModel)
tModel = Dropout(0.2)(tModel)
tModel = Dense(1, activation="sigmoid")(tModel)
model = Model(inputs=bModel.input, outputs=tModel)
5º Step
Determine the hyperparameters and compile the model
The hyperparameters, in particular, the learning rate (“INIT_LR”), the epochs (“EPOCHS”) and the batch size (“BS”) were determined.
Subsequently, the Adam optimization function (“opt”) was defined, the model was compiled considering the loss function “binary_crossentropy” and as a metric for evaluating the results, accuracy was considered.
INIT_LR = 1e-3
EPOCHS = 50
BS = 8
for layer in bModel.layers:
layer.trainable = False
opt = Adam(lr=INIT_LR)
model.compile(loss="binary_crossentropy", optimizer=opt,
metrics=["accuracy"])
6º Step
Train the model
From the command below, the model was trained, leaving 10% of the images for validation. The information was saved in variable “x” and the model was saved on the computer as “modeloc_1.hdf5”.
x = model.fit(X_train, Y_train, batch_size=BS,validation_split=0.1, epochs=EPOCHS)
model.save("/content/drive/My Drive/Python/COVID/model/modeloc_1.hdf5")
7º Step
Observe the accuracy of the model and the loss function
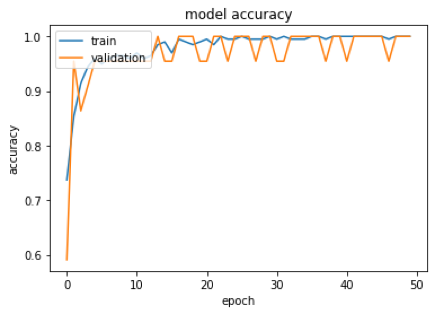
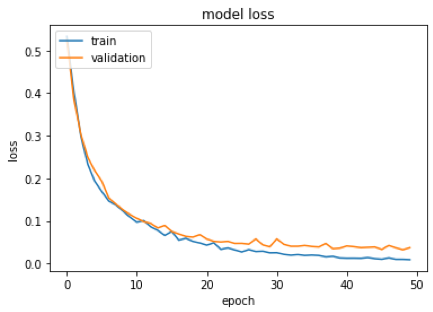
We built a graph to analyze the accuracy history of training data and model validation. We also built a graph that computes the network error in relation to the training and validation data. They point out that, apparently, there was no overfitting, since the training and validation lines approached.
In addition, it is noted that the model’s accuracy was 98%. That is, the model hit 98% of the images used in the test.
plt.plot(x.history['accuracy'])
plt.plot(x.history['val_accuracy'])
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'validation'], loc='upper left')
plt.show()
plt.plot(x.history['loss'])
plt.plot(x.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'validation'], loc='upper left')
plt.show()
model.evaluate(X_test,Y_test)
2/2 [==============================] - 1s 342ms/step - loss: 0.0345 - accuracy: 0.9818
[0.03453369066119194, 0.9818181991577148]
8º Step
Observe which images the model hit
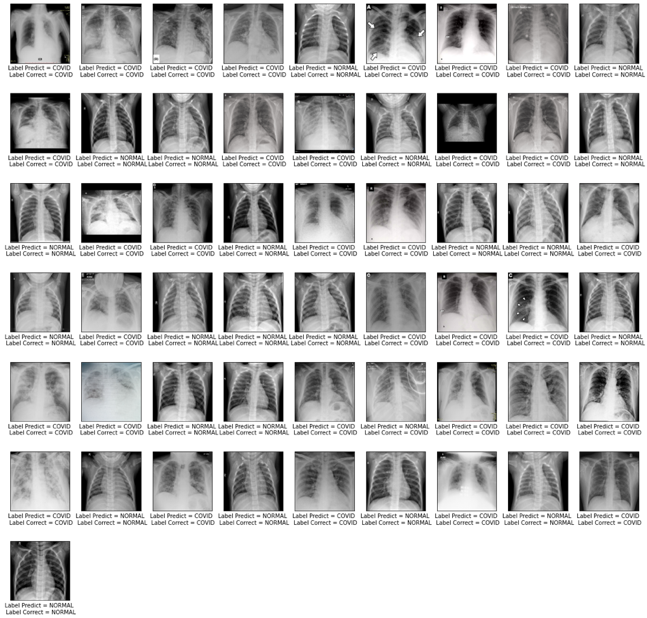
From the image below it is possible to see the images that the model got right. The “Labels” (Label Predict and Label Correct) that have the same name indicate that the model has correctly predicted. Example: Label Predict = COVID and Label Correct = COVID. In this sense, it is possible to observe that the model hit 54 out of 55 total images.
In addition, the figure was saved as modelo_1.pdf on the computer.
plt.figure(figsize=(20,20))
i = 0
for i,image in enumerate(X_test):
plt.subplot(7,9,i+1)
plt.xticks([])
plt.yticks([])
plt.grid(False)
plt.imshow(image, cmap=plt.cm.binary)
img = np.expand_dims(X_test[i],axis = 0)
x_pred = model.predict(img)[0]
x0 = x_pred[0]
if x0 > 0.5:
label = "COVID"
else:
label = "NORMAL"
if Y_test[i] == 1:
label_test = "COVID"
else:
label_test = "NORMAL"
plt.xlabel(f"Label Predict = {label} \n Label Correct = {label_test}")
i += 1
plt.savefig('/content/drive/My Drive/Python/COVID/model/modelo_1.pdf')
9º Step
Build a confusion matrix
The code below creates a confusion matrix with the model data.
The model only missed one classification among the 55 images used for the test, presenting an accuracy of 98%. The confusion matrix shows that, among the total images, 58% (n = 32) represent true positives, 40% (n = 22) true negatives, 1.8% (n = 1) false negatives and 0% (n = 0) false positives.
ypredict = model.predict(X_test)
ypredictc = []
for value in ypredict:
x0 = value [0]
# x1 = value [1]
if x0 > 0.5:
ypredictc.append(1)
else:
ypredictc.append(0)
resultado = np.array(ypredictc)
x = confusion_matrix(y_true=Y_test,y_pred=resultado)
x = x/X_test.shape[0]
y = pd.DataFrame(x,index = ["NORMAL","COVID"],columns=["NORMAL","COVID"])
plt.figure(figsize = (10,7))
fig = sn.heatmap(y, annot=True,cmap="Greens").get_figure()
fig.savefig("plot.jpg")

Conclusion on model 1: From the preliminary results it is possible to notice that the model has a high accuracy to classify the lungs normal and with COVID-19. The next model will train with images of lungs that have other infections, in order to obtain a model capable of differentiating COVID-19 from other infections.
Note: the results are not clinical, but exploratory. However, with the improvement of the models, they can bring benefits to confront COVID-19.
Bibliography
COHEN, Joseph; MORRISON, Paul; DAO, Lan. COVID-19 Image Data Collection. arXiv:2003.11597, 2020.
KERMANY, Daniel; ZHANG, Kang; GOLDBAUM, Michael. Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification. Mendeley Data, v.2, 2018. Disponível em: http://dx.doi.org/10.17632/rscbjbr9sj.2